Protein Dynamics and Function
Rather than being static entities, proteins are capable of adopting numerousrelated conformations that are dependent on the protein’s environment. Protein flexibility and dynamics play important roles in carrying out their functions. [1] [2] [3] Instead of a single static conformation, proteins will populate ensembles of often quite similar structures and transitions between these states occur on a variety of time scales, from the picosecond to the millisecond. It seems, therefore, that determining the tertiary or quaternary structure of a protein is not an end in and of itself, but a stepping stone to full understanding of protein behaviour.
Instead of there being a single static structure at the end of the protein folding pathway, the protein folding pathway is a dynamic equilibrium between a number of related conformations, and the interchange between these structures can occur on a number of time scales. The kinetics and thermodynamics of the system will ensure that the majority of molecules will inhabit a conformation close to the global minimum, but exchange between these related conformers will occur at biological temperatures.
It is now generally accepted that primary structure determines tertiary structure, which in turn determines dynamics and hence function, but as yet no direct measurement has been recorded that establishes the exact relationship between dynamics, structure and function. This is due, in part, to the fact that protein behaviour occurs across numerous time-scales, from the picosecond to the second: changes in secondary structure happen over nanoseconds to milliseconds; the movement of a large side chain occurs on the order of tens of picoseconds and rapid changes in local structure and environment as a result solvent interactions can happen in just a few picoseconds. It is difficult to carry out a single experiment that probes all these time-scales simultaneously.
However, much work has been conducted to flesh out the picture of protein dynamics on a picosecond time scale. A range of tools is herefore required in order to investigate the full range of protein motions on various time-scales. In the picosecond range, [00 Inbox/Sleeping/PhD/Two-Dimensional Infrared Spectroscopy|Two-Dimensional Infrared Spectroscopy] may be used to probe the behaviour of proteins in solution. Experimental 2DIR measurements of proteins may be complemented by calculated spectra derived from molecular dynamics simulations. A combination of the two may allow the nature of the relationship between structure, dynamics and function on a picosecond to nanosecond time-scale to investigated in greater detail.
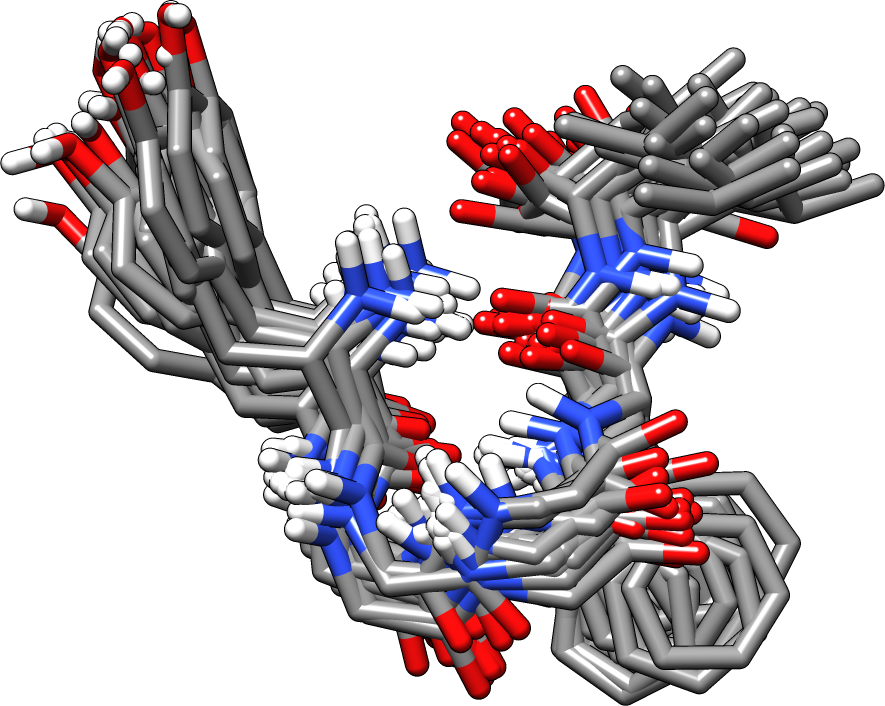
An ensemble of 20 closely related structures for Leu-enkephalin in vacuum, taken from an MD simulation.
Planted: Friday, 24 February 2023
Last tended: Monday, 23 June 2025
C. Micheletti. Comparing proteins by their internal dynamics: Exploring structure function relationships beyond static structural alignments. Phys. Life Rev., 10:1–26, 2013. ↩︎
P.C. Biggin. Protein dynamics — a moving target: Comment on “Comparing proteins by their internal dynamics: Exploring structurefunction relationships beyond static structural alignments” by C. Micheletti . Phys. Life Rev., 10:27–28, 2013. ↩︎
S. Hammes-Schiffer and J. Klinman. Emerging Concepts about the Role of Protein Motion in Enzyme Catalysis. Acc. Chem. Res., 48:899–899, 2015. ↩︎